L7 | CHATS
‘everything data’ series
The L7 and SciBite Partnership Enables AI Implementation Through Ontology-backed Data Unification and Harmonization
by Brigitte Ganter, Ph.D. | posted on July 24, 2024
This is the second installment of our series of blog entries that will focus on “everything data.”
Laboratory and manufacturing processing data can comprise numerous structured or unstructured data sets which are often not aligned to any standard terminology and therefore heterogeneous. Reusing laboratory and manufacturing processing data for secondary process insight generation has gained importance and is seen as the only way to understand process bottlenecks, identify cost sinks, and detect process inefficiencies. Collecting these data can be a nightmare, especially if not all are in the same structure or in the same system, a common challenge across the therapeutics industry. This is where L7 Informatics with its Unified Platform L7|ESP®, comes into play. As summarized in our last AI post, Building the Foundation for Artificial Intelligence Applications, the L7|ESP platform, with its Workflow Orchestration and Data Contextualization, is built with an architecture for digitalization. L7|ESP not only integrates all common data information management systems, including L7 LIMS, L7 Notebooks, and L7 MES together with other process-oriented applications, but is also open and flexible to integrate with existing management systems and even works with legacy systems, providing a unified view of all laboratory and business processes across the value chain, thereby increasing transparency of the product life cycle and its information flow, while ensuring operational efficiency.
L7|ESP harmonizes and structures data at the point of capture and can also be extended with data trending and charting tools for real-time operational and scientific insight.
Download the L7|ESP data sheet to learn more about L7|ESP.
Ontologies Are Key to Extracting Maximal Value from Laboratory and Manufacturing Process Data and Laying the Foundation for AI
To date, many process data analytics projects fail or do not deliver the expected outcome, mostly due to impaired access to all the relevant data or low-quality, unstructured data. Ideally, data would be aggregated into a single system and structured and aligned to common standards. The industry is not at that point, and unstructured data and siloed systems are the norm.
For process data analytics projects to be successful, the data feeding these analytics needs to be well structured and use standardized terminology. A data-centric approach, ensuring data is fit for purpose and aligned to standard terminology, is required. The only effective way of achieving this is via an ontology-backed approach. Ontologies support the development and management of a consensus view of knowledge in a domain, including hierarchical relationships between entities and the mapping of synonyms. Aligning data to ontologies ensures consistency of terminology and interoperability of data, supporting robust analytics and efficient extraction of insights. Data aggregation and alignment to ontologies make information more transparent to both internal (e.g., data consumers) and external stakeholders (e.g., audiences such as regulators) and are crucial to achieving business efficiency. Ontologies, which are networks of human-generated, machine-readable descriptions of knowledge, are a critical tool for making data FAIR (Findable, Accessible, Interoperable, and Reusable), and as such, reusable for business applications.
A semantic model (i.e., an ontology) is required to organize data that shows the basic meaning of data items and the relationships between them. This can help with knowledge sharing, reasoning, and understanding of concepts.
Ontologies Solve the Pharmaceutical Industry’s Massive Issue of Information Transparency and Interoperability by Making Data FAIR
Rich public ontologies are addressing the life sciences’ unique challenge in relation to the heterogeneity and complexity of language and terminology. They organize and represent concepts, properties, and their relationships. Public ontologies are a structured representation of knowledge agreed upon by experts and represent a consensus view of a particular field, e.g., genes, drugs, or assays. Each concept within a public ontology contains a unique identifier and by adopting these public identifiers, data becomes interoperable within the organization and between different organizations. These individual identifiers are persistent and remain unchanged, even if the preferred term or synonyms associated with them change. This provides a future-proof solution to data interoperability.
Ontologies classify and explain entities
Ontologies classify and explain entities (i.e., things or classes) via a common vocabulary. In other words, they formally conceptualize the content of a specific domain, e.g., “rare disease” or “laboratory and manufacturing” domains in the therapeutics industry. In the world of laboratory processes, this means a “DNA sequencer” is a type of “equipment,” with subtypes being a “benchtop sequencer” or “production-scale sequencer.” The classes may have contextual definitions that a human can use to understand what the class contains, as well as synonyms and relationships to other classes. For example, there are classes of sequencers of type “Illumina sequencer” for “short-read sequencing,” or in other words, they are “short-read sequencer,” while sequencers of type “PacBio sequencer” are for “long-read sequencing”, hence they are “long-read sequencer” (Side note: Illumina also now offers a complete long-read technology and PacBio a short-read sequencer). Following this classification, not every sequencer is a “short-read sequencer” but a “sequencer.” Ontologies help define the relationships between them.
An ontology-backed approach is important when it comes to laboratory and manufacturing processes and extracting insights from the data collected to improve laboratory and business processes.
This is further demonstrated when it comes to downstream processing of sequence data. Accurate mapping of sequencing reads to a reference genome or transcriptome relies on the use of high-quality alignment and mapping tools. Since the products generated using one of those “sequencer” (i.e., “short-read sequencer” versus “long-read sequencer”) are mapped and processed differently, read length-specific software, or in other words, “sequence mapping software,” is required. To map the product generated by a “short-read sequencer,” a “short-read sequence mapping tool” is required. One popular “short-read sequence mapping tool” is the BWA package, which includes three options of algorithms: BWA-backtrack, BWA-SW, and BWA-MEM. Whereas for long-read sequences, a “long-read sequence mapping tool” is required, of which examples are Minimap2 and LRA. To complicate matters, Minimap2 can also be used to map short-read sequences. Which brings me to my point: all these details are important to refine and optimize the process, and therefore, these details need to be tracked and aligned to a structured ontology.
L7 Informatics and SciBite Partnership for Effective AI Implementation Through Ontology-backed Data Harmonization
Understanding the value that ontologies provide for data unification and interoperability, L7 Informatics has partnered with SciBite to support the use of SciBite’s CENtree ontology management platform with L7|ESP. CENtree simplifies the implementation of public ontologies and standards and allows its users to seamlessly merge public ontologies with proprietary terminology, facilitating the development, management, and use of ontologies in data models within the L7|ESP platform.
Thus, data can be aligned to public standards at the point of capture in L7|ESP, using ontology-backed picklists, and following FAIR data principles.
Furthermore, existing CENtree users can bring their enterprise ontologies from CENtree into L7|ESP through the integration between these tools (i.e., the integration with L7|MASTER) as depicted in Figure 1, aligning terminology across their business and serving as the single source of truth for terminology. Non-CENtree users are also able to benefit via an embedded version of CENtree available as a module within L7|ESP (not shown).
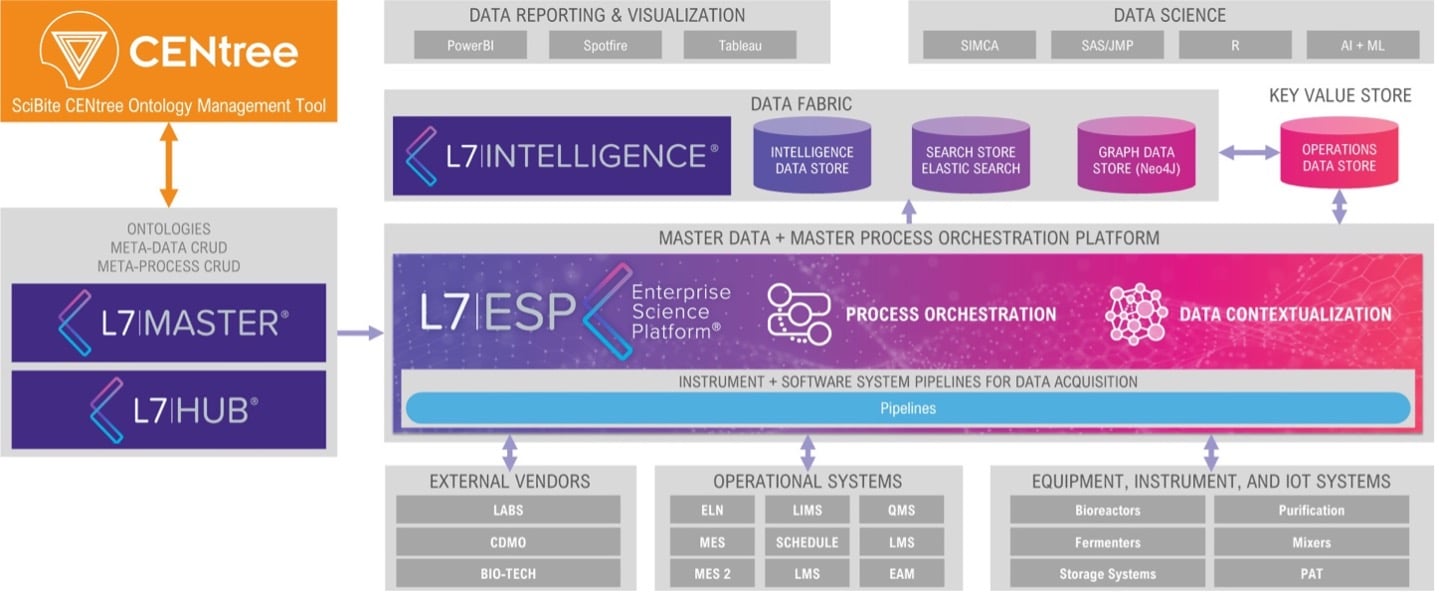
Figure 1: L7|ESP integration with CENtree.
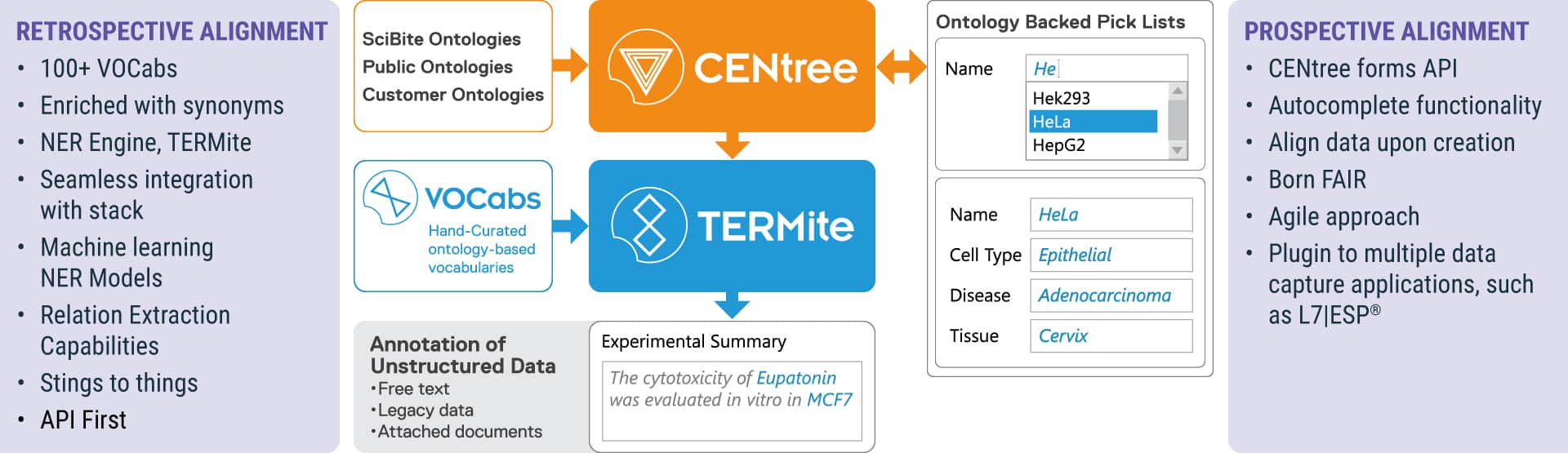
Figure 2: Prospective capture workflow using ontology-backed pick lists using the SciBite platform of applications.
The L7 Informatics / SciBite partner integration enables customers of both companies to leverage L7|ESP data models, public ontologies, or internal data models and ontologies managed in CENtree.
About SciBite’s CENtree and TERMite
Ontology-backed picklists allow prospective capture of structured data in alignment with enterprise-wide terminology. The prospective capture of data via ontology-backed pick lists and the retrospective alignment of legacy and unstructured data to ontologies using TERMite is shown in Figure 2. Through alignment with public ontologies, data is aligned more broadly with agreed upon terminology within the scientific community. However, legacy data and unstructured data are often captured in the form of free-text experimental write-ups and attached documents, such as Word, PowerPoint, or PDF documents attached to experiments. This type of data is not amenable to a picklist that has already been captured, and this presents a challenge. Specifically, the data captured is, in many instances, not using common terminology and lacks structure which limits its usage.
Using a process called Named Entity Recognition (NER), these same ontologies developed and managed in CENtree can also be used to standardize and structure legacy data. Ontologies can be exported from CENtree in a format that is compatible with SciBite’s TERMite NER tool. TERMite processes this unstructured text, identifying concepts within the text and assigning public identifiers and additional metadata to the legacy data. The enriched data can then be presented in a unified format in L7|ESP, enabling its usage for AI applications. SciBite also offers over 100 VOCabs covering a broad range of concepts in the life sciences that are based on public ontologies but are optimized for NER and for extracting entities from unstructured data. This optimization is done by SciBite’s ontology team and includes automated and manual synonym expansion as well as creating rules that allow the context in which an entity occurs in the text to be considered, improving the accuracy of entity recognition.
CENtree is SciBite’s enterprise ontology management platform (Ontology Manager), which supports the development and deployment of ontologies at enterprise scale and the integration into downstream applications via an API-first architecture.
TERMite is SciBite’s named entity recognition tool (a Text Analysis Engine), allowing legacy and unstructured data to be retrospectively aligned to ontologies.
VOCabs are SciBite’s Named Entity Recognition optimized vocabularies based on public ontologies (Premade Expert Ontologies). In conjunction with TERMite they can support the harmonization and structuring of unstructured or semi-structured data in alignment to public ontologies.
Key Benefits of the L7 Informatics-SciBite Partnership
- Existing CENtree users can connect their respective platforms to bring ontologies and the terminology managed in CENtree to L7|ESP.
- Non-CENtree users can take advantage of SciBite’s ontology management CENtree module within L7|ESP.
- Merge public ontologies and standards with your internal terminology via application ontologies, which then can be pushed to L7|ESP as data models.
- Streamline the development and ongoing management of ontologies in L7|ESP.
- Prospectively align data to ontologies at the point of capture in L7|ESP.
- Retrospectively align legacy and unstructured data to ontologies via SciBite’s TERMite NER tool.
- Take advantage of AI-ready, harmonized data aligned to standards.
Combining L7|ESP with SciBite’s CENtree ontology management solution provides L7|ESP users with a robust, ontology-backed data capture and unification tool that supports complex life science workflows and product supply chains. Not only can it be implemented quickly, but it also provides a powerful data management tool aligned with FAIR data principles.
